Fitting XShooter data with xtool¶
In [1]:
from xtool.data import XShooterData, Order
from xtool.model import OrderModel, VirtualPixelWavelength, GenericBackground, MoffatTrace, SlopedMoffatTrace
from collections import OrderedDict
from scipy import sparse
from scipy import optimize
import numpy as np
Reading XShooter data¶
In [2]:
xd = XShooterData('xtool_ds/')
In [3]:
current_order = xd[17]
current_order.enable_flags_as_mask() # important to update the data to include the bad pixels from the pipeline
Generating a virtual pixel table for “Wavelength”-pixels¶
In [4]:
virt_pix = VirtualPixelWavelength.from_order(current_order, wavelength_sampling=0.03)
pixel_table = virt_pix()
Initializing the two Models¶
In [5]:
background_mdl = GenericBackground(pixel_table, virt_pix.wavelength_pixels)
trace_mdl = SlopedMoffatTrace(pixel_table, virt_pix.wavelength_pixels)
In [6]:
order_model = OrderModel([background_mdl, trace_mdl])
Show fittable parameters¶
In [7]:
order_model
Out[7]:
<OrderModel(background_level=[ 0. 0. 2023. ..., 2008. 2008. 2008.], amplitude=[ nan nan nan ..., nan nan nan], trace_pos=0.0, trace_slope=0.0, sigma=1.0, beta=1.5 [f])>
Initializing the fitter¶
In [8]:
from xtool.model.fitters import Fitter
In [9]:
fitter = Fitter(order_model, current_order)
Differential Evolution fit¶
Differential evolution is a very thorough but slow processs. It does not use starting values but moves within the bounds of the parameters.
In [31]:
order_model.beta.fixed = False
dresult = fitter.fit_differential_evolution(disp=True)
differential_evolution step 1: f(x)= 4.22785e+06
differential_evolution step 2: f(x)= 4.22785e+06
differential_evolution step 3: f(x)= 2.95083e+06
differential_evolution step 4: f(x)= 2.95083e+06
differential_evolution step 5: f(x)= 2.26008e+06
differential_evolution step 6: f(x)= 2.26008e+06
differential_evolution step 7: f(x)= 2.26008e+06
differential_evolution step 8: f(x)= 2.26008e+06
differential_evolution step 9: f(x)= 2.06608e+06
differential_evolution step 10: f(x)= 2.06231e+06
differential_evolution step 11: f(x)= 2.00095e+06
differential_evolution step 12: f(x)= 1.96445e+06
differential_evolution step 13: f(x)= 1.96445e+06
differential_evolution step 14: f(x)= 1.96445e+06
differential_evolution step 15: f(x)= 1.89723e+06
differential_evolution step 16: f(x)= 1.89723e+06
differential_evolution step 17: f(x)= 1.89723e+06
differential_evolution step 18: f(x)= 1.89723e+06
differential_evolution step 19: f(x)= 1.89654e+06
differential_evolution step 20: f(x)= 1.89654e+06
differential_evolution step 21: f(x)= 1.89654e+06
Fit finished in 1475.61700892 s
In [32]:
dresult
Out[32]:
fun: 1896544.4149950482
message: 'Optimization terminated successfully.'
nfev: 1425
nit: 21
success: True
x: array([-2.44188896, -0.26832235, 0.19794569, 2.0046534 ])
Nelder Mead / Simplex¶
This one is much faster algorithm that will use starting values, but is unbounded
In [10]:
order_model.trace_pos = -2
order_model.sigma = 0.1
order_model.trace_slope = 0.
order_model.beta.fixed = False
order_model.beta = 2.0
result = fitter.fit_scipy_minimize('Nelder-Mead')
/Users/wkerzend/anaconda3/envs/xtool/lib/python2.7/site-packages/scipy/optimize/_minimize.py:394: RuntimeWarning: Method Nelder-Mead cannot handle constraints nor bounds.
RuntimeWarning)
Fit finished in 851.559898853 s
/Users/wkerzend/anaconda3/envs/xtool/lib/python2.7/site-packages/scipy/sparse/linalg/dsolve/linsolve.py:243: SparseEfficiencyWarning: splu requires CSC matrix format
warn('splu requires CSC matrix format', SparseEfficiencyWarning)
/Users/wkerzend/anaconda3/envs/xtool/lib/python2.7/site-packages/scipy/sparse/linalg/dsolve/linsolve.py:161: SparseEfficiencyWarning: spsolve is more efficient when sparse b is in the CSC matrix format
'is in the CSC matrix format', SparseEfficiencyWarning)
In [15]:
spec = order_model.model_list[1].to_spectrum()
In [19]:
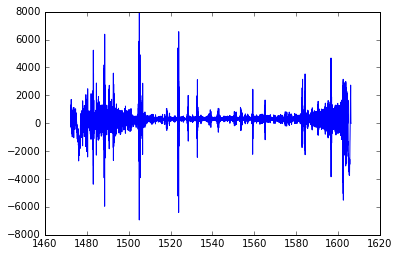
%pylab inline
plot(spec.wavelength, spec.flux)
Populating the interactive namespace from numpy and matplotlib
Out[19]:
[<matplotlib.lines.Line2D at 0x1231a7a50>]
Plotting in DS9¶
In [11]:
import pyds9
In [12]:
d = pyds9.DS9()
In [14]:
order_model.trace_pos = result.x[0]
order_model.trace_slope = result.x[1]
order_model.sigma = result.x[2]
order_model.beta = result.x[3]
model = order_model.evaluate_to_frame(current_order, trace_pos=order_model.trace_pos,
trace_slope=order_model.trace_slope, sigma=order_model.sigma,
beta=order_model.beta.value)
d.set('frame 1')
d.set_np2arr(current_order.data.filled())
d.set('frame 2')
d.set_np2arr(model.filled())
d.set('frame 3')
d.set_np2arr((current_order.data.filled() - model.filled()) / current_order.uncertainty.filled())
Out[14]:
1